
Author: Prof. Dr. Kathrin Schumann
Genetic manipulation of human primary cells has been largely impossible until recently, but advances in genome engineering methods offer new opportunities. Genome engineering of eukaryotic cells with CRISPR/Cas9 has been first described in 2012 and has since then revolutionized this research field (Jinek et al. 2013). It is widely accepted that T-cell genome engineering holds great promise for cell-based therapies for cancer, HIV and autoimmune diseases, but genetic manipulation of human T cells has been challenging.
So far, our knowledge about this unique cell type is largely based on data generated in transgenic mice or with RNAi approaches in human T cells which exhibit significant off-target effects. We developed a robust CRISPR/Cas9 technology based on Cas9 ribonucleoproteins (Cas9 RNPs), recombinant Cas9 protein complexed with chemically synthesized gRNAs, that enables both knock-out and knock-in genome editing in primary human T cells to overcome this hurdle (Schumann and Lin et al. PNAS 2015).
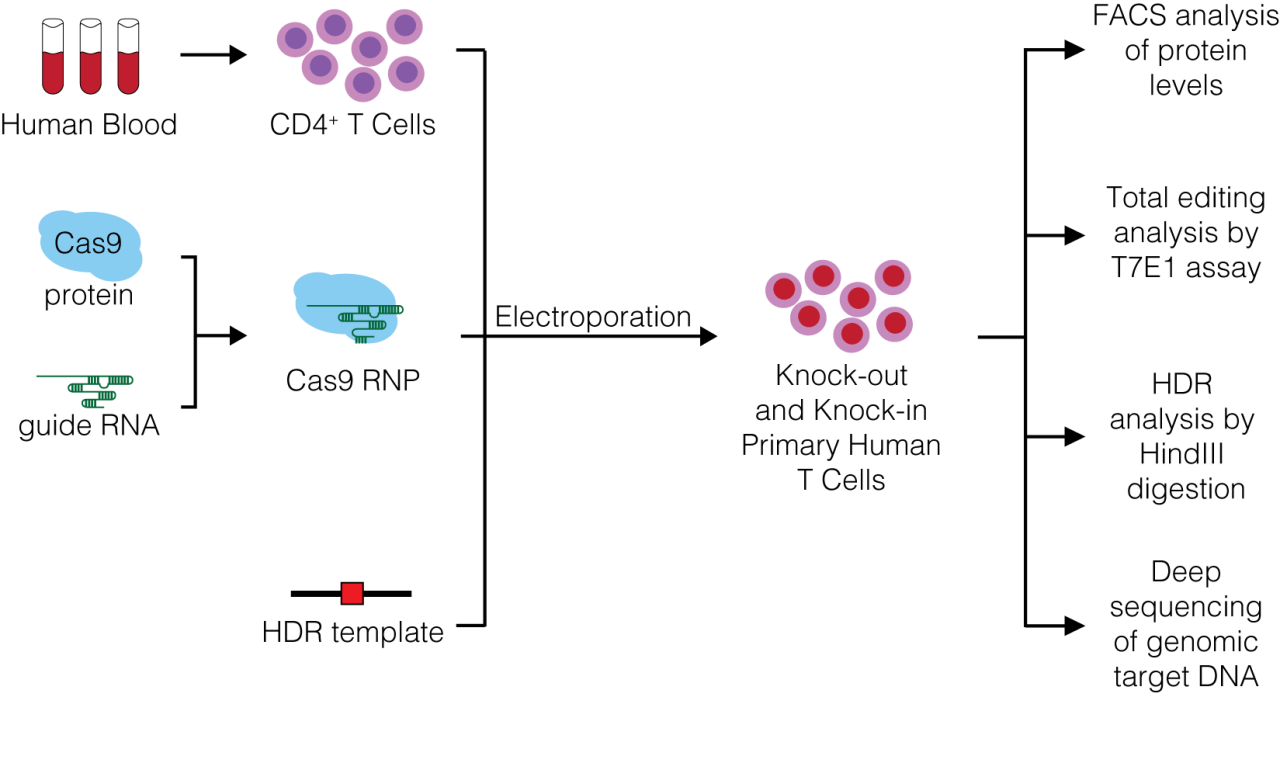
Figure 1: Generation of knock-out and knock-in primary human T cells using Cas9 ribonukleoproteins (Cas9 RNPs).
We expanded the CRISPR/Cas9 technique to a variety of genes associate with HIV (Hultquist & Schumann et al. Cell Reports 2016), cancer (Rupp & Schumann et al. Scientific Reports 2017) and autoimmune diseases.
CRISPR screens in primary human T cells
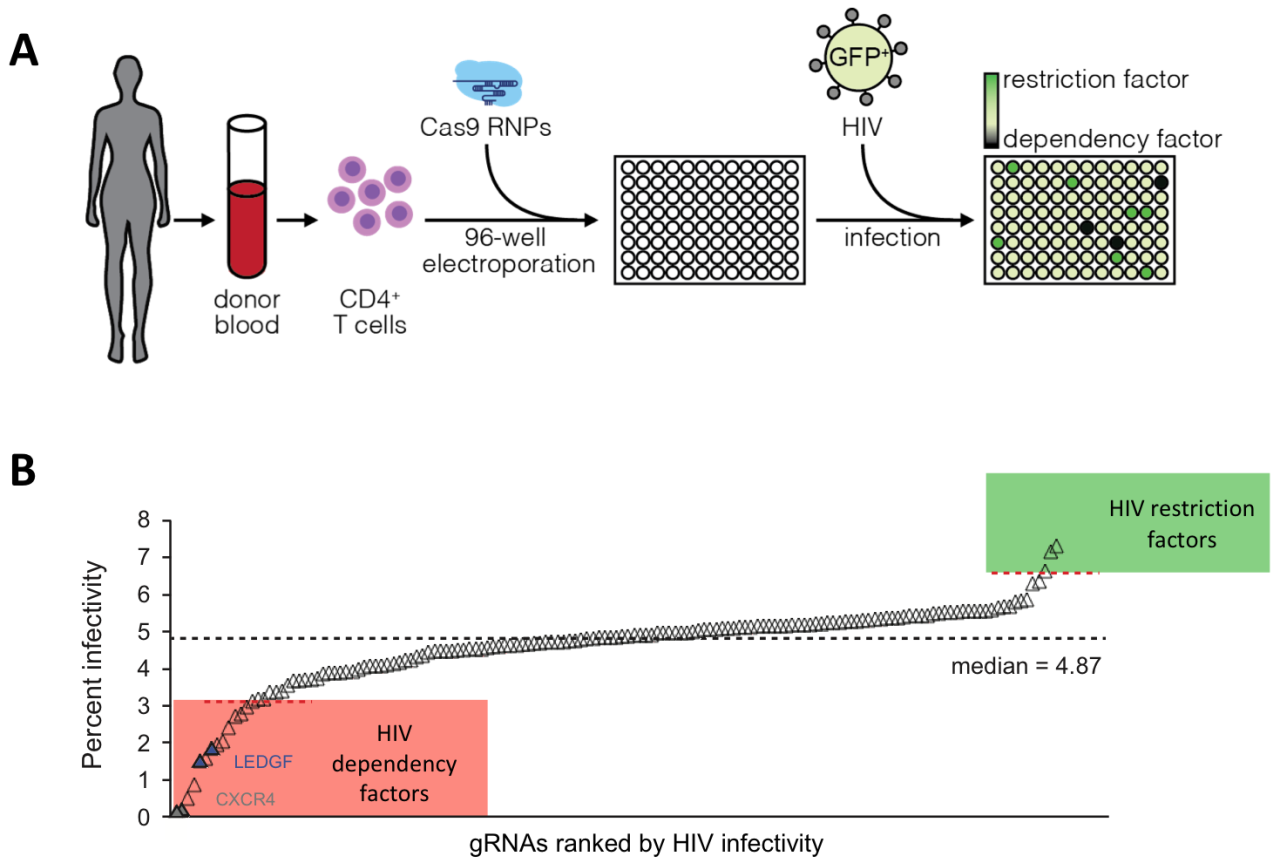
We developed a medium-throughput platform to systematically ablate HIV host factors in primary human T cells in a rapid, arrayed fashion using Cas9 RNPs in a 96 well format, which has the potential to accelerate target validation for pharmaceutical and cellular therapies. We systematically screened for known and predicted HIV integrase interaction partners in T cells and could identify novel dependency and restriction factors (Hultquist & Schumann et al. Cell Reports 2016).
We apply this arrayed platform now to analyse the stability and the transcriptional patterns of different human T cell subset. We have a special interest in applying these technologies to human regulatory T cells.
Dissecting transcriptional regulation in human regulatory T cells
Regulatory T cells (Tregs) play a fundamental role in maintaining immune tolerance by suppressing autoreactive effector T cells. Tregs with pro-inflammatory features have been described in different context and with a variety of phenotypes. Murine “ex-Tregs” downregulate their master transcription factor Foxp3 and in humans and mice with autoimmune conditions Tregs with changed cytokine profile could be detected. However, the processes that lead to these heterogeneous phenotypes are incompletely understood. Methods to stabilize Tregs for the treatment of autoimmune diseases or actively destabilize Tregs to ablate tolerogenic effects on the tumor microenvironment have great therapeutic potential. To identify these potential targets for therapy we perform arrayed and pooled CRISPR screens to systematically ablate transcription factors in human Tregs.
We are interested in dissecting the gene networks regulated by individual transcription factors in human T cells by performing arrayed and pooled CRISRP screens. Besides that, we want to develop novel ex vivo models to functionally validate human CRISPR-engineered T cells.
With these insights we hope to engineer advanced cell therapies against autoimmunity or cancer in the future.
*dieser Inhalt ist nur auf Englisch verfügbar*